Introduction
In this part we’re going to explore some popular Python tools for data science.
NumPy
Numerical Python, is the standard package for computation and array operations.
Low-level functions are written in C, therefore, very fast.
Basic operations in NumPy
We usually import as:
import numpy as npNumPy arrays can be constructed with the following:
A = np.array([69, 420, 1337, 42])A NumPy array is defined by:
- The number of dimensions it has (
ndim) - The number of elements it has (
size) - Its
shape(number of elements along an axis) - Its
dtype
A = np.array([69, 420, 1337, 42])# A.ndim = 1# A.size = 4# A.shape = (4, )# A.dtype = int64We can either specify the dtype or let NumPy automatically assign it.
Different ways to create NumPy arrays that we usually use:
arangeconstruct numbers from a range.zeroscreates an array full of zeros, with any shape.onescreates an array full of ones, with any shape.eyecreates the identity array.fullcreates an array filled with a specific element/array.
We can reshape our arrays and matrix, the new shape must have equal size (product of the shape).
A = np.array([69, 420, 1337, 42])A.reshape(2, 2)'''[[69, 420], [1337, 42]]'''This does not make a copy, this returns a view (basically a pointer).
We can transpose our arrays and matrix as well:
A = np.array([69, 420, 1337, 42])A.reshape(2, 2).T'''[[69, 1337], [420, 42]]'''We can change the dtype with the .astype() function:
A = np.array([69, 420, 1337, 42])A.astype(np.float32)'''[69.0, 420.0, 1337.0, 42.0]'''We access elements in NumPy arrays with usual bracket notation:
A = np.array([69, 420, 1337, 42])A[0]'''69'''NumPy supports Python style slicing:
A = np.array([69, 420, 1337, 42])# A[1:-1] = [420, 1337]# A[::2] = [69, 1337]NumPy supports all basic math operations:
A = np.array([69, 420, 1337, 42])# A + 1 = [70, 421, 1338, 43]There are a lot more functions to explore :)
Matplotlib
Matplotlib is the standard way of plotting figures and graphs in Python. Used for ploting any kind of plot you can think of, scatter plots, line plots, contour plots, etc.
It comes with multiple APIs, but the standard one for Python is the pyplot API. Let’s take a look at a practical example.
Anscombe’s quartet
Anscombe’s quartet is a small dataset that shows the importance of graphing your data when dealing with statistics.
To understand we’ll graph this dataset for ourselves:
import numpy as npimport matplotlib.pyplot as pltFirstly, let’s import numpy for the data processing and matplot for the plotting.
anscombe_data = np.array([10.0, 8.04, 10.0, 9.14, 10.0, 7.46, 8.0, 6.58,8.0, 6.95, 8.0, 8.14, 8.0, 6.77, 8.0, 5.76,13.0, 7.58, 13.0, 8.74, 13.0, 12.74, 8.0, 7.71,9.0, 8.81, 9.0, 8.77, 9.0, 7.11, 8.0, 8.84,11.0, 8.33, 11.0, 9.26, 11.0, 7.81, 8.0, 8.47,14.0, 9.96, 14.0, 8.10, 14.0, 8.84, 8.0, 7.04,6.0, 7.24, 6.0, 6.13, 6.0, 6.08, 8.0, 5.25,4.0, 4.26, 4.0, 3.10, 4.0, 5.39, 19.0, 12.50,12.0, 10.84, 12.0, 9.13, 12.0, 8.15, 8.0, 5.56,7.0, 4.82, 7.0, 7.26, 7.0, 6.42, 8.0, 7.91,5.0, 5.68, 5.0, 4.74, 5.0, 5.73, 8.0, 6.89])Let’s reshape our NumPy array so it is a bit more readble and how it looks in Wikipedia.
anscombe_data = anscombe_data.reshape(11, 4, 2).transpose(1, 0, 2)'''[[[10. 8.04] [ 8. 6.95] [13. 7.58] [ 9. 8.81] [11. 8.33] [14. 9.96] [ 6. 7.24] [ 4. 4.26] [12. 10.84] [ 7. 4.82] [ 5. 5.68]]
[[10. 9.14] [ 8. 8.14] [13. 8.74] [ 9. 8.77] [11. 9.26] [14. 8.1 ] [ 6. 6.13] [ 4. 3.1 ] [12. 9.13] [ 7. 7.26] [ 5. 4.74]]
[[10. 7.46] [ 8. 6.77] [13. 12.74] [ 9. 7.11] [11. 7.81] [14. 8.84] [ 6. 6.08] [ 4. 5.39] [12. 8.15] [ 7. 6.42] [ 5. 5.73]]
[[ 8. 6.58] [ 8. 5.76] [ 8. 7.71] [ 8. 8.84] [ 8. 8.47] [ 8. 7.04] [ 8. 5.25] [19. 12.5 ] [ 8. 5.56] [ 8. 7.91] [ 8. 6.89]]]'''Let’s break it up into the four indviual datasets:
anscombe = {'I': anscombe_data[0, :, :], 'II': anscombe_data[1, :, :], 'III': anscombe_data[2, :, :], 'IV': anscombe_data[3, :, :]}Before we plot the actual graphs, let’s take a loook at the mean, standard devation and variance for all four of the datasets.
for key, value in anscombe.items(): print(key) print('Mean: ', np.mean(value, axis=0)) print('Standard deviation: ', np.std(value, ddof=1, axis=0)) print('Variance: ', np.var(value, axis=0)) print('Correlation coefficient: ', np.corrcoef( value[:, 0], value[:, 1])[0, 1]) print()'''IMean: [9. 7.50090909]Standard deviation: [3.31662479 2.03156814]Variance: [10. 3.75206281]Correlation coefficient: 0.81642051634484
IIMean: [9. 7.50090909]Standard deviation: [3.31662479 2.03165674]Variance: [10. 3.75239008]Correlation coefficient: 0.8162365060002428
IIIMean: [9. 7.5]Standard deviation: [3.31662479 2.0304236 ]Variance: [10. 3.74783636]Correlation coefficient: 0.8162867394895984
IVMean: [9. 7.50090909]Standard deviation: [3.31662479 2.03057851]Variance: [10. 3.74840826]Correlation coefficient: 0.8165214368885028'''So, from a purerly statistical view, we would think that all these datasets should look somewhat similar, right?
Let’s plot them and see. Let’s plot them as scatter plots:
for key, value in anscombe.items(): plt.scatter(value[:, 0], value[:, 1]) plt.show()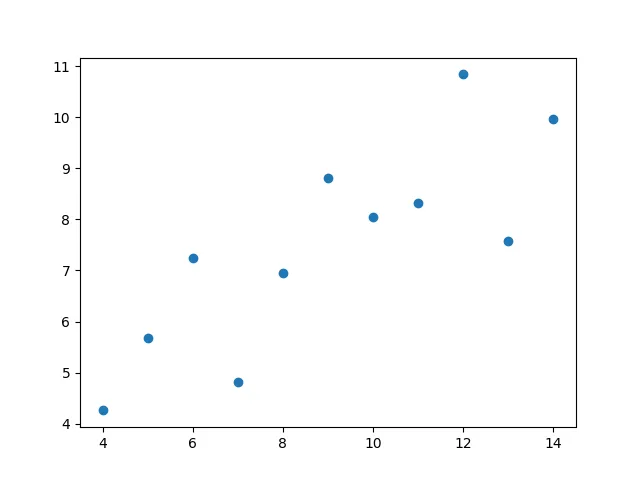
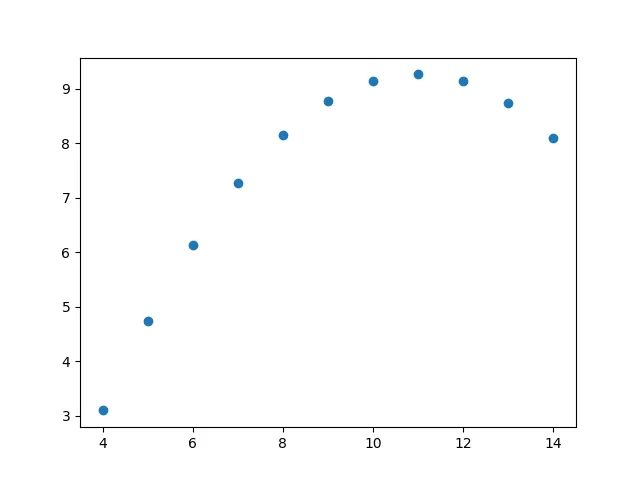

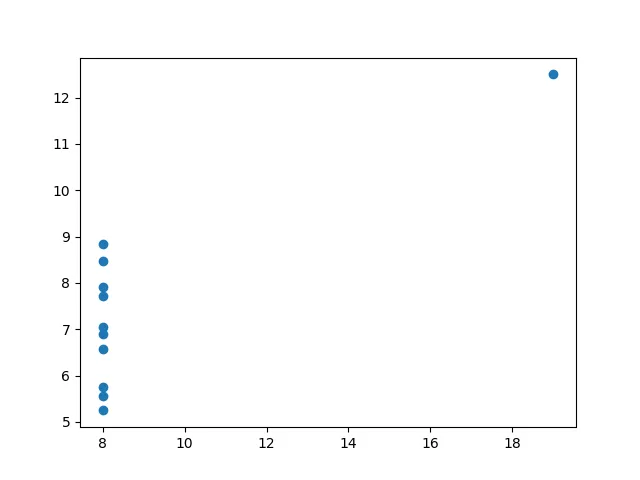
Let’s plot them all together:
fig, axs = plt.subplots(2, 2)for (ax, (key, value)) in zip(axs.ravel(), anscombe.items()): ax.scatter(value[:, 0], value[:, 1]) ax.set_xlabel('$x$') ax.set_ylabel('$y$') ax.set_title(key)
fig.tight_layout()plt.show()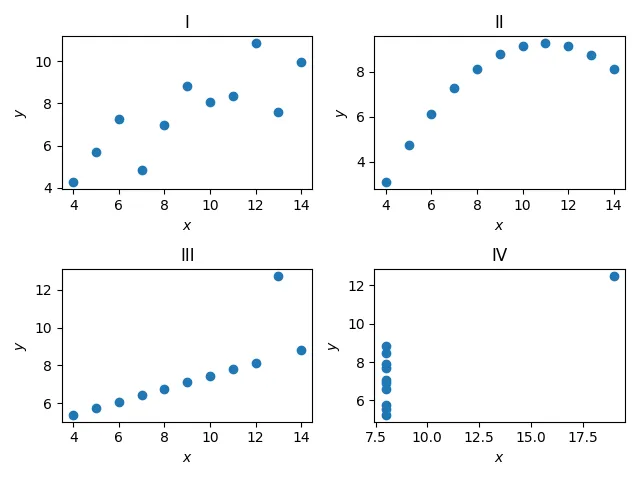
So, we can see that these datasets, in reality, differ a lot from each other, but seem to have equivalent statistical properties. We’ll look into more statistics later on :).
Pandas
The last library we’ll cover is the data analysis library Pandas. When dealing with large datasets, we’ll usually want to use common convient functions to read, write and manipulate data fast.
Pandas can make use of NumPy as a backend for some data.
Before jumping into a practial example we’re we’ll use Pandas, let’s first understand how we’ll represent data.
Wide and long format
When we want to represent data, we have two options:
- In wide format, the data is indexed by first column where the values do not repeat.
- Different variables for observations are placed on different columns.
- In long format, a column indexes the different kinds of observations and another column contains the respective values.
Converting data to wide format is called pivoting.
Converting data to long format is unpivoting or melting.
'''Wide format
Person Age Weight Height0 Bob 32 168 1801 Alice 24 150 1752 Steve 64 144 165
Long format
Person Attribute Value0 Bob Age 321 Bob Weight 1682 Alice Age 243 Alice Weight 1504 Steve Age 645 Steve Weight 144'''Palmer penguins
We’ll use the palmer penguins data set.
Let’s import Pandas, note that we don’t import NumPy here!
import pandas as pdPandas offers a great selection of reading methods for different file formats.
df = pd.read_csv('penguins_size.csv')print(df)''' species island culmen_length_mm culmen_depth_mm flipper_length_mm body_mass_g sex0 Adelie Torgersen 39.1 18.7 181.0 3750.0 MALE1 Adelie Torgersen 39.5 17.4 186.0 3800.0 FEMALE2 Adelie Torgersen 40.3 18.0 195.0 3250.0 FEMALE3 Adelie Torgersen NaN NaN NaN NaN NaN4 Adelie Torgersen 36.7 19.3 193.0 3450.0 FEMALE.. ... ... ... ... ... ... ...339 Gentoo Biscoe NaN NaN NaN NaN NaN340 Gentoo Biscoe 46.8 14.3 215.0 4850.0 FEMALE341 Gentoo Biscoe 50.4 15.7 222.0 5750.0 MALE342 Gentoo Biscoe 45.2 14.8 212.0 5200.0 FEMALE343 Gentoo Biscoe 49.9 16.1 213.0 5400.0 MALE'''To get an overview we can use the .describe() function.
print(df.describe())''' culmen_length_mm culmen_depth_mm flipper_length_mm body_mass_gcount 342.000000 342.000000 342.000000 342.000000mean 43.921930 17.151170 200.915205 4201.754386std 5.459584 1.974793 14.061714 801.954536min 32.100000 13.100000 172.000000 2700.00000025% 39.225000 15.600000 190.000000 3550.00000050% 44.450000 17.300000 197.000000 4050.00000075% 48.500000 18.700000 213.000000 4750.000000max 59.600000 21.500000 231.000000 6300.00000'''To select a column we can do:
print(df['species'])'''0 Adelie1 Adelie2 Adelie3 Adelie4 Adelie ...339 Gentoo340 Gentoo341 Gentoo342 Gentoo343 GentooName: species, Length: 344, dtype: object'''We can also select data with .iloc and .loc. iloc is index based.
print(df.iloc[0])'''species Adelieisland Torgersenculmen_length_mm 39.1culmen_depth_mm 18.7flipper_length_mm 181.0body_mass_g 3750.0sex MALEName: 0, dtype: object'''Where as .loc is string based, in this case both iloc and loc yields the same answer since we group the data with index.
In other datasets we could have a string based index.
We can easily filter and choose the right data with:
print(df.loc[df['culmen_length_mm'] < 40])''' species island culmen_length_mm culmen_depth_mm flipper_length_mm body_mass_g sex0 Adelie Torgersen 39.1 18.7 181.0 3750.0 MALE1 Adelie Torgersen 39.5 17.4 186.0 3800.0 FEMALE4 Adelie Torgersen 36.7 19.3 193.0 3450.0 FEMALE5 Adelie Torgersen 39.3 20.6 190.0 3650.0 MALE6 Adelie Torgersen 38.9 17.8 181.0 3625.0 FEMALE.. ... ... ... ... ... ... ...146 Adelie Dream 39.2 18.6 190.0 4250.0 MALE147 Adelie Dream 36.6 18.4 184.0 3475.0 FEMALE148 Adelie Dream 36.0 17.8 195.0 3450.0 FEMALE149 Adelie Dream 37.8 18.1 193.0 3750.0 MALE150 Adelie Dream 36.0 17.1 187.0 3700.0 FEMALE
[100 rows x 7 columns]'''We can have chained boolean expressions with & for AND, | for OR, and ~ for NOT.
print(df.loc[(df['culmen_length_mm'] < 60) & (df['species'] == 'Gentoo')])''' species island culmen_length_mm culmen_depth_mm flipper_length_mm body_mass_g sex220 Gentoo Biscoe 46.1 13.2 211.0 4500.0 FEMALE221 Gentoo Biscoe 50.0 16.3 230.0 5700.0 MALE222 Gentoo Biscoe 48.7 14.1 210.0 4450.0 FEMALE223 Gentoo Biscoe 50.0 15.2 218.0 5700.0 MALE224 Gentoo Biscoe 47.6 14.5 215.0 5400.0 MALE.. ... ... ... ... ... ... ...338 Gentoo Biscoe 47.2 13.7 214.0 4925.0 FEMALE340 Gentoo Biscoe 46.8 14.3 215.0 4850.0 FEMALE341 Gentoo Biscoe 50.4 15.7 222.0 5750.0 MALE342 Gentoo Biscoe 45.2 14.8 212.0 5200.0 FEMALE343 Gentoo Biscoe 49.9 16.1 213.0 5400.0 MALE
[123 rows x 7 columns]'''Very often it is useful to sort/index the raw data based on some metric, let’s group the numerical data by the species and take the mean:
print(df.groupby('species')[['culmen_length_mm', 'culmen_depth_mm', 'flipper_length_mm', 'body_mass_g']].mean())''' culmen_length_mm culmen_depth_mm flipper_length_mm body_mass_gspeciesAdelie 38.791391 18.346358 189.953642 3700.662252Chinstrap 48.833824 18.420588 195.823529 3733.088235Gentoo 47.504878 14.982114 217.186992 5076.016260'''Let’s format this into long format:
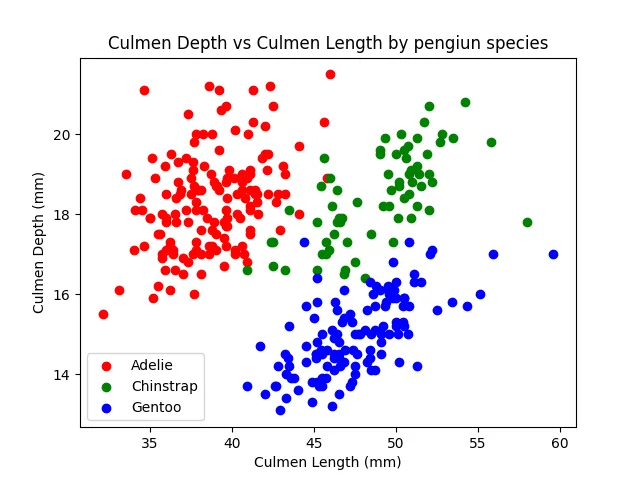
df2 = df.groupby('species')[['culmen_length_mm', 'culmen_depth_mm', 'flipper_length_mm', 'body_mass_g']].mean()print(df2.reset_index().melt(id_vars=['species'], var_name='measurement', value_name='value'))''' species measurement value0 Adelie culmen_length_mm 38.7913911 Chinstrap culmen_length_mm 48.8338242 Gentoo culmen_length_mm 47.5048783 Adelie culmen_depth_mm 18.3463584 Chinstrap culmen_depth_mm 18.4205885 Gentoo culmen_depth_mm 14.9821146 Adelie flipper_length_mm 189.9536427 Chinstrap flipper_length_mm 195.8235298 Gentoo flipper_length_mm 217.1869929 Adelie body_mass_g 3700.66225210 Chinstrap body_mass_g 3733.08823511 Gentoo body_mass_g 5076.016260'''Lastly, let’s make a simple plot that shows the culmen depth vs culmen length for all the species in one scatter plot.
import matplotlib.pyplot as pltimport numpy as npcolors = ['red', 'green', 'blue']species_to_num = {v: k for (k, v) in enumerate(df['species'].unique())}
for species, group in df.groupby('species'): plt.scatter(group['culmen_length_mm'], group['culmen_depth_mm'], color=colors[species_to_num[species]], label=species) plt.xlabel('Culmen Length (mm)') plt.ylabel('Culmen Depth (mm)') plt.title('Culmen Depth vs Culmen Length by pengiun species') plt.legend()
plt.show()